
Project area C: Theory and computational analysis
C1 Evolution of gene expression
| Project leaders: | Johannes Berg, Institut für Theoretische Physik, Universität zu Köln Michael Lässig, Institut für Theoretische Physik, Universität zu Köln |
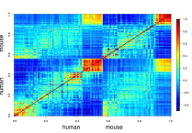
Gene expression levels are important molecular traits of an organism. They are determined by the processes of transcription, translation, and degradation of proteins and mRNA. We will infer selection pressures on gene expression and on the underlying processes, using new transcriptomic and proteomic data in yeast. For example, we will ask how selection drives the correlation of the expression levels of the genes in a cellular pathway. The long-term goal is to achieve a comprehensive map of the evolutionary forces shaping gene expression levels in yeast.
C2 Adaptive evolution of microbial and viral systems
| Project leader: | Michael Lässig, Institut für Theoretische Physik, Universität zu Köln |
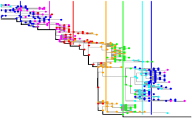
Adaptive evolution is driven by changes in an organism's environment. We will develop quantitative models of adaptive evolution in microbial and viral systems, focusing on two systems: (a) Evolution of Escherichia coli in a mouse gut ecosystem, in collaboration with project B13. (b) Evolution of the human influenza A virus (the picture shows a tree of influenza strains over a period of seven years). We will build predictive models of influenza evolution, which establish a link to the epidemiology of this system and can inform vaccine selection.
C3 Epistasis, recombination and predictability in adaptive evolution
| Project leader: | Joachim Krug, Institut für Theoretische Physik, Universität zu Köln Arjan de Visser, Laboratory of Genetics, Wageningen University |
The project addresses fundamental aspects of evolutionary adaptation through a combination of population genetic theory and experimental evolution. Using the antibiotic-resistance enzyme TEM-1 β-lactamase, we study interactions between mutations increasing resistance, and construct models that explain the mapping from genotype to fitness through intermediate phenotypes. Based on such empirical fitness landscapes, we investigate the factors that determine the repeatability of evolution, including population size and recombination rate. Our long-term goal is to understand the roles of contingency and determinism in adaptation, and how they limit the predictability of evolution.
C4 Evolutionary innovations – a case study on chromatin insulators
| Project leader: | Thomas Wiehe, Institute for Genetics, Universität zu Köln |

Chromatin insulators play a fundamental role in the spatio-temporal expression of genes and in adaptive evolution of phenotypic traits. Interestingly, the insulator protein complement of different lineages, e.g. of arthropods and vertebrates, shows considerable variation. To understand the causes and consequences of this variation, we will integrate macro- and micro-evolutionary analyses of genetic diversity and divergence, of gene expression profiles, of DNA- and protein-interaction networks. In addition, we will use a bioinformatics approach to search for new insulator proteins. Results are expected to reveal how these important chromatin regulators contribute to animal evolution.
C7 The role of horizontal gene transfer in life style changes and metabolic adaptations
| Project leader: | Martin Lercher, Institut für Informatik, Heinrich-Heine-Universität Düsseldorf |
Anecdotal evidence shows that horizontal gene transfers, i.e., the acquisition of individual genes from other species, have boosted the metabolic and extremophilic capabilities of bacteria, archaea, and even eukaryotes. To examine this issue systematically, we will combine comparative genomics with ecological data and detailed metabolic network analyses. We will test for correlations of phenotypic data with gene transfers as well as gene duplications, thereby increasing our understanding of how these processes contributed to environmental adaptation.
C9 The evolution of repetitive DNA and its methylation
| Project leader: Achim Tresch, Botanisches Institut, Universität zu Köln |
Alu retrotransposons are the most abundant class of selfish, short DNA elements that proliferate by RNA-mediated reverse transcription. We will measure the evolutionary fitness of Alu sequence variants in terms of their reintegration rates into the genome. Our goal is to identify the molecular causes of evolutionary fitness, such as germline transcription, stability of Alu RNA, and its capability to reintegrate into the genome. By combining phylogenetic data with DNA methylation- , transcriptomics-, and Chromatin immunoprecipitation data, we will quantify the influence of sequence variation on these causes.
Past projects
C6 Control and evolution of somite pattern formation during early development (2006-2009)
| Project leader: | Ulrich Gerland, Ludwig-Maximilians-Universität München |
Vertebrate somitogenesis is being studied in a diverse set of animals, and its understanding has reached a level where theoretical modeling can provide important insights. Here, we focus on an evolutionary transition in the oscillatory expression pattern of the genes involved in the ‘somitogenesis clock’. Such a transition appears to have occurred between Zebrafish and Medaka. We will use theoretical modeling to study the phenotypic variability of the regulatory network controlling somitogenesis, and will perform experiments to characterize the effects of the regulatory evolution in key clock elements.


