
Project area A: Evolution between species
A1 The emergence of Toll signaling as major component of the dorsoventral patterning network during insect evolution
| Project leader: | Siegfried Roth, Institut für Entwicklungsbiologie, Universität zu Köln |
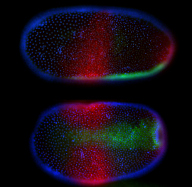
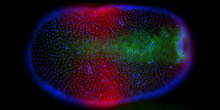
In the course of insect evolution a signaling pathway (the Toll pathway), which had an ancestral function in immunity, acquired a new role for the formation of the dorsoventral body axis of the embryo. To investigate how this occurred at the molecular level we will use genomic and functional approaches to study axis formation in the bug Oncopeltus (representing the more basal hemimetabolous insects) and the beetle Tribolium (representing the more advanced holometabolous insects).
A5 The evolution of seed plant stem cell niches: the homeodomain perspective
| Project leader: | Wolfgang Werr, Institut für Entwicklungsbiologie, Universität zu Köln |
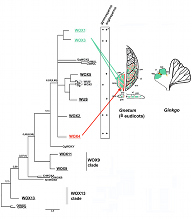
Plant development depends on stem cell niches, which in the Arabidopsis shoot and root meristems are promoted by WUSCHEL (WUS) and WOX5, respectively. Both are members of the WUSCHEL related homeobox or WOX gene family, which has a monophyletic origin in green algae and evolved via gene amplifications from basal to higher plants. Most recent additions were member in the WUS-clade, which were amplified prior seed plants. Synapomorphic traits serve specific stem cell niches through gymno- and angiosperms i.e. WOX2 in the apical embryo domain, WOX3 in marginal or plate meristems and WOX4 in the vascular cambium. The recruitment of WUSCHEL to the organizing center of the shoot apical meristem (SAM) and WOX5 to the quiescent center of the root involved a duplication followed by sub-functionalisation unique to angiosperms. The project addresses functional changes from the ancestral WOX13 lineage towards more recent family additions and has a specific emphasis on the evolutions of the DNA-binding homeodomain (HD). We use macro-evolutionary information provided by lineage-specific signatures of WOX-HD family members, X-ray diffraction data obtained for the WUS-HD and covariance/protein sector analysis to guide functional studies with regard to the HD function in the SAM stem cell niche of the Arabidopsis model.
A9 Macro-evolution of a gene-regulatory network in Brassicaceae
| Project leader: | Martin Hülskamp, Botanisches Institut, Universität zu Köln |
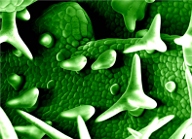
Trichome and root hair density are important adaptive traits that are regulated by partially overlapping gene networks in Arabidopsis. We propose to study the evolution of these networks in Brassicaceae focusing on three aspects: (1) impact of gene duplications, (2) changes in network gene expression and readout, (3) constraints on one pathway controlling two traits. We combine a functional analysis in Arabis alpina with molecular studies of additional Brassicaceae species. Our goal is to understand compensatory and adaptive changes at the network level, and how these changes lead to new phenotypes.
A10 Evolution of transcriptional regulation during the divergence of annual and perennial plant life histories
| Project leader: | George Coupland, Max Planck Institut für Pflanzenzüchtungsforschung, Köln |

This project studies evolution of the plant life cycle. Annual plants live for less than a year and flower only once, whereas perennials live for many years and flower each year. Annuals have evolved from perennial progenitors many times. Here we propose to study the diversification of these traits using crosses between annual and perennial sister species in the Brassicaceae. We will utilize this material to explore the genetic basis of species-specific patterns in the regulation and function of the MADS box transcription factor PERPETUAL FLOWERING 1.
A12 The changing roles of Hox3 genes in insect evolution
| Project leader: Kristen Panfilio, Institut für Entwicklungsbiologie, Universität zu Köln |
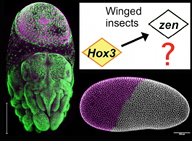
What is the basis for novelty during evolution? In this project we will address the origin of the extraembryonic membranes in insects, an adaptive innovation that protects the developing embryo. This innovation correlates with functional changes in the transcription factor Hox3/Zen. Through detailed molecular and functional analyses in two insect species and broader genomic comparisons, we will elucidate (1) the original role of Zen within this new morphological context, and (2) how subsequent changes across the insects have led to further diversity and complexity.
A13 An interdisciplinary approach for understanding diversification of leaf form
| Project leader: Miltos Tsiantis, Max Planck Institut für Pflanzenzüchtungsforschung |
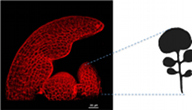
A key challenge in biology is to understand how diversity in organismal form is generated. We developed Cardamine hirsuta - a relative of the model plant Arabidopsis thaliana - into a powerful system for studying morphological evolution. Here we aim to understand the morphogenetic paths leading to the strikingly different geometries of the two species: simple leaves in A. thaliana and dissected leaves with leaflets in C. hirsuta. To achieve this we will use a combination of genetics, interspecies gene transfers, advanced imaging and computational modeling. The project will be instrumental in producing predictive models of a leaf shape, a trait that evolves in close correspondence with the environment suggesting it is of high adaptive significance.
Past projects
A2 Evolution of the regulatory interactions of the segmentation gene network in insects (2006-2008)
| Project leader: | Diethard Tautz, Max-Planck-Institut für Evolutionsbiologie, Plön |
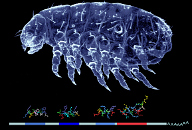
The segmentation gene hierarchy in Drosophila is one of the best studied regulatory networks to date and there is rapidly accumulating sequence information from insect genomes that can be used for comparisons. We have used the established genetic model systems D. melanogaster and Tribolium castaneum to unravel conserved and divergent principles of the segmentation gene hierarchy, as well as its evolutionary dynamics. We have been able to identify a novel segmentation gene in Tribolium, have done a functional comparative analysis of the hunchback and hairy gene in both species and have contributed to the comparative analysis of the whole Tribolium genome.
A4 Analysis of a new type of MADS-box gene that is involved in the evolution of land plant gametophytes (2006-2009)
| Project leader: | Thomas Münster, Max-Planck-Institut für Planzenzüchtungsforschung, Köln |
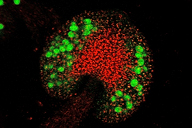
Using a combinatorial approach (analysis of phylogeny, molecular mechanisms, networking, function) we have been able to reconstruct the evolutionary history of the gametophyte-specific MIKC* MADS-box genes in land plants. Moreover, the respective MIKC* protein networking and the evolution of their dimerization capability could be identified in detail in representatives of informative land plant lineages, including the characterization of DNA binding sites of a number of MIKC* protein complexes using a random oligonucleotide binding site selection method. In addition, putative target genes have been characterized with the help of gene chip technology in MIKC* mutants in Arabidopsis.
Among the genes with an altered expression level in MIKC* mutant background the TIR1 receptor for the plant hormone auxin could be identified as the major target gene candidate of the MIKC* genes in Arabidopsis.
A6 Evolution of the reproductive mode in nematodes (2006-2009)
| Project leader: | Einhard Schierenberg, Zoologisches Institut, Universität zu Köln |
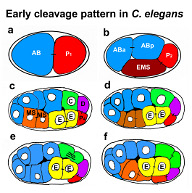
We are interested in the evolutionary transition from a bisexual to parthenogenetic mode of reproduction. For this we investigate developmental aspects of parthenogenetic nematodes with special reference to meiosis, establishment of polarity and cellular pattern formation and compare these to the hermaphroditic C. elegans as a standard and other nematodes with bisexual reproduction. Besides analysis of cellular modifications during oocyte and embryo development, we cloned genes that are homologous to ones essential for for oocyt-to-embryo transition in C. elegans. Via RNA interference we attempt to knock down these genes in C.elegans to elucidate their function. In addition, we study genome-wide gene expression in mutants where certain steps of germ cell development are impaired using DNA microarrays.


