
Project area B: Evolution within species
B3 Co-evolution of pathogen virulence and host defense
| Project leader: | Jonathan C. Howard, Institut für Genetik, Universität zu Köln / Instituto Gulbenkian de Ciência |
Polymorphic virulence proteins of the intracellular protozoal parasite, Toxoplasma gondii, interact with polymorphic resistance proteins (IRG proteins) of the mouse. We will analyse the genetic and phenotypic diversity of mouse IRG proteins and known Toxoplasma virulence factors from infected and uninfected wild mice. The results of these experiments will inform a co-evolutionary model that describes the ecological fitness interactions between host-pathogen genotype combinations and predicts the resulting evolutionary dynamics.
B10 Temperature modulation of plant immune responsiveness and fitness
| Project leader: | Jane E. Parker, Max-Planck-Institut für Züchtungsforschung, Köln |
Mechanisms of plant environmental adaptation are poorly understood. Here we explore evolutionary and molecular processes underlying selection of immunity genes in Arabidopsis local populations. We will measure defence, growth and fitness traits associated with locally adapted immune receptor allelic forms and the influence of temperature on the trade-off between their induced resistance pathways and fitness.
B11 Cost and benefit of bacterial transformation
| Project leader: | Berenike Maier, Institut für Theoretische Physik und Biozentrum, Universität zu Köln |

Bacterial transformation enables bacteria to exchange genetic information and can therefore speed up adaptive evolution. It is unclear however, under which conditions the benefit of transformation outweighs its cost. The goal of this project will be to experimentally quantify the costs and benefits of transformation in Neisseria gonorrhoeae. In particular we will test the influence of reversible mutations in genes that generate costs in transformable strains. In the long run, this project will contribute to our understanding of how recombination has evolved and is stably maintained.
B12 The adaptive significance of small-effect mutations in yeast and plants
| Project leader: Andreas Beyer, Institut für Genetik, Universität zu Köln |
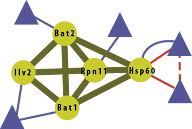
Mutations often only have small effects on molecular traits such as mRNA or protein levels. Yet, many such mutations might together have evolutionary relevant effects on physiological parameters. Detecting adaptation through small effect mutations is a statistically challenging problem. We propose using innovative computational methods that combine molecular data at the level of networks to overcome the statistical difficulties associated with small effects. This approach is expected to reveal new insights into how small effect mutations translate to molecular and pathway changes that ultimately affect cellular and tissue traits.
Associated project B13 Adaptation of Escherichia coli to a complex ecosystem
| Project leader: | Isabel Gordo, Instituto Gulbenkian de Ciência |
Microbial evolution in complex ecosystems is a subject of growing interest. Laboratory mice models constitute a compromise between a truly natural environment and in vitro setups such as chemostats. We will study bacterial adaptation in the ecosystem comprising the mouse gut and focus on how mutation rate and effective population size affect genomic and phenotypic patterns of adaptive evolution. We will test predictions of theoretical models of adaptive evolution in collaboration with project C2.
Past projects
B1 Characterization of potential adaptive trait loci in house mouse populations (2006-2008)
| Project leader: | Diethard Tautz, Max-Planck-Institut für Evolutionsbiologie, Plön |

Using genomic scans for signatures of positive selection in wild populations of the house mouse, we have identified a number of candidate genes that may have contributed to very recent adaptations. Some of these loci were further studied based on population genetical parameters and functional approaches. A special focus was given on changes of regulation, as well as on genes expressed in testis. A systematic comparative study on the role of positive selection in shaping expression divergence was conducted between subspecies of the mouse. A novel gene was identified that has evolved within the subgenus Mus and that appears to play a role in sperm motility.
B2 The role of regulatory changes in speciation prosesses of the house mouse (2006-2008)
| Project leader: | Bettina Harr, Max-Planck-Institut für Evolutionsbiologie, Plön |
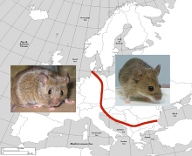
Under the biological species concept, the generation of reproductive isolation is synonymous with speciation. Understanding the genes that contribute to reproductive isolation is essential to understanding speciation, but isolating such genes has proven very difficult. In this study, we propose to use natural hybrid zones and expression profiling to identify genes that are miss-regulated in hybrids between two house mouse subspecies, M. m. domesticus and M. m. musculus. This approach will give an insight into both the number and kind of genes involved in reproductive isolation.
B4 Identification of Genes involved in Local Selection and Genetic Adaptation in Humans (2006-2009)
| Project leaders: | Peter Nürnberg, Cologne Center for Genomics and Institute for Genetics, Universität zu Köln Manfred Kayser, Erasmus University Medical Center, Rotterdam |

The genetic process under which humans have adapted to new environmental factors during their migration history remains largely unknown. We use high-density genome-wide SNP scans to pinpoint genomic regions that are likely to have been shaped by local positive selection in Pacific Islanders. Applying a battery of test statistics we are searching for partial and complete selective sweeps and estimate their statistical significance by means of simulations considering ascertainment bias as well as population histories. Candidate regions are scrutinized for genes and other functional elements that might have been the selection targets. As we want to test the thrifty gene hypothesis we are focussing on genes involved in obesity and obesity-related diseases such as diabetes type 2. The most convincing candidates will be followed-up by detailed analyses including functional studies. Verified functional variants are supposed to explain the process of recent adaptation to varying environmental sources on the molecular level.
B5 The adaptive value of pleiotropic variation: miR824 and the MADS-Box Gene Network (2006-2013)
| Project leaders: | Juliette de Meaux, Institut für Evolution und Biodiversität, Westfälische Wilhelms-Universität Münster Maarten Koorneef, Max-Planck-Institut für Züchtungsforschung, Köln |
The current challenge of evolutionary theory is to identify the molecular mechanism by which populations respond to natural selection and adapt to new environments. This project aims at elucidating the ecological cause and functional consequences of variation segregating at high frequency in the miR824 gene, encoding a microRNA known to control one member of a complex transcription factor network. For this, molecular constructs will be developed to identify the phenotypic consequences of miR824 polymorphism. Theoretical models will be constructed to explain the maintenance of polymorphism at this locus and the outcome of the model will be tested with field assays of fitness under ecologically relevant conditions.
B7 Evolutionary adaptation of Daphnia to protease inhibitors in cyanobacteria (2010-2013)
| Project leader: | Eric von Elert, Zoologisches Institut, Universität zu Köln |
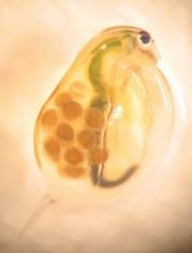
For D. magna, the most important herbivores in lakes, digestive trypsins and chymotrypsins have been shown to be the targets of dietary cyanobacterial protease inhibitors, and the respective protease genes have been identified. In a D. magna population it will be investigated if the seasonal appearance of protease inhibitors affects the frequency of protease alleles and what the molecular mechanisms of these micro-evolutionary changes are. In clonal cultures of D. magna the tolerance of protease inhibitors will be linked to mutations in and expression levels of protease genes. In a correlative approach, copy number variation in protease families will be examined for their potential source of adaptive mutations.


